Analysis and visualization software for massive next-generation sequencing data sets
Presented by: Riku Katainen, PhD student, Aaltonen lab, Genome-Scale Biology Program
http://research.med.helsinki.fi/gsb/aaltonen/
Due to the massive availability of next- and third generation sequencing (NGS) data, the processing and analysis now constitute a serious challenge for research in life sciences. I will introduce a highly efficient and user friendly analysis and visualization software tool (BasePlayer), which is designed for huge NGS data sets. BasePlayer offers comparative variant analyses not only in genes but also in intergenic regions by integrating regulatory annotation, transcription factor (TF) binding sites and motifs.
I am going to show basic functions of the program in real time with an actual cancer data. BasePlayer is still under development and will be published soon, so I’ll be happy to hear your suggestions and comments.
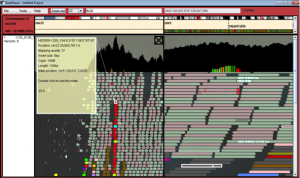
Screenshot of somatic mutations in APC gene and annotation in Base Player
Screenshot of split screen view at TTC28 LINE1-Element in Base Player