I’m Pinja Perkkiö, an aspiring microbiologist and a HiLIFE scholarship trainee in Reetta Satokari’s lab (University of Helsinki, Faculty of Medicine)! I’ve just finished my bachelor’s degree and I will start my studies in the master’s programme of Microbiology and Microbial biotechnology next autumn. This summer, I will be diving deep into the world of gut microbiota as I research the microbiota restoring potential of NGPs in intestinal inflammation and dysbiosis.
How antibiotics disturb our gut microbiota
The gut microbiota is defined as a community of microbes, e.g. bacteria, fungi, viruses and archaea residing within the gut. The microbiota is responsible for many important functions, such as breaking down complex carbohydrates and protecting the gut from infections. When a person takes antibiotics, it not only kills the bacteria causing the disease, but also the beneficial ones as antibiotics aren’t strain-specific. This allows potentially pathogenic bacteria to grow in larger numbers – the situation is called dysbiosis, meaning that the microbial community composition has been altered in a harmful way.
Luckily, we have a tight layer of cells – the epithelium – that lines the walls of the gut and is covered by two layers of mucus. The mucus combined with the epithelium acts as a barrier between our intestines and the rest of our body, preventing intestinal pathogens from causing too much harm. Normally, it is very difficult for microbes to cross the mucus layer as it traps microbes and is loaded with antibacterial substances. In addition, the cells of the epithelium are tightly attached to each other, preventing any further movement of microbes. There are also lots of immune cells embedded in gut tissues that are ready for action, should a pathogen try to cause an infection.
However, as dysbiosis leads to the growth of pathogenic bacteria, our immune cells (neutrophils, NK cells and macrophages, for example) activate inflammatory reactions. While these reactions are necessary for the elimination of pathogens, they also damage gut tissue. Pathogenic bacteria also activate the secretion of inflammatory molecules, cytokines, in a type of gut epithelial cell called enterocyte, which leads to the epithelium getting “leaky” – this means that microbes can cross the epithelium more easily and cause further inflammation in gut tissue. These bacteria can not only cause nasty infections, but they may also travel deeper in gut tissue or even to other sites in the body, increasing the risk of certain cancers (Bastos et al., 2023; Kouzu et al., 2022)!
Next-generation probiotics against dysbiosis
To fight antibiotic-induced dysbiosis and reduce the inflammation and leaky gut syndrome caused by it, researchers have turned their attention to potential new bacterial strains that could function as next-generation probiotics. They are defined as “live microorganisms identified on the basis of comparative microbiota analyses that, when administered in adequate amounts, confer a health benefit on the host” (Martín R, Langella P., 2019) – in simpler terms, microbes that can potentially help boost our health. Traditional probiotics, such as Bifidobacteria and Lactobacilli, have shown great potential in restoring a healthy microbial community in the gut after taking antibiotics, thus preventing dysbiosis and infections. They have also been shown to inhibit the production of inflammatory molecules in both immune cells and epithelium and boost the production of anti-inflammatory molecules.
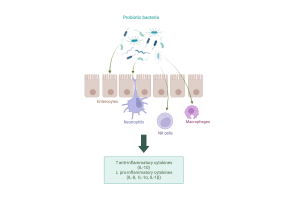
Probiotics exert anti-inflammatory effects on several cell types. Created with Biorender.com.
There’s still lots of research to be done to bring next-generation probiotics into wide use. More information is needed on the different strains of potential probiotic bacteria and their specific effects on both the immune system and the gut epithelium. This is where my research project comes in!
The project I’m collaborating on focuses on a mix of 14 different anaerobic bacterial strains that have the potential to restore the gut microbiota after antibiotic use and prevent or reduce inflammation. The next-generation probiotic mix was originally tested in an in vitro mucosal-simulator of the human gastrointestinal tract (M-SHIME(R)) model. A natural gut microbiota was obtained from the stool sample of a healthy human donor, after which it could grow. Then, the “gut” was exposed to antibiotics to induce dysbiosis. After the antibiotic treatment, one system was treated with the probiotic mixture and the other one with a negative control. Finally, samples from probiotic- and control-treated microbiotas were taken. Bacteria present on these will be studied to check the impact of the antibiotic intake in the microbiota and compare its possible recovery after the probiotic treatment. The production of short-chained and branched fatty acids, lactate and ammonia at different time points will also be measured since their levels indicate how much beneficial bacteria there are compared to harmful ones.
To study how the anaerobic isolates might affect intestinal inflammation, I will culture two different intestinal epithelial cell lines: Caco-2 and HT-29. These cell lines were chosen because they are well-characterized and resemble enterocytes on the gut epithelium. Then, I will expose the HT-29 cell line to either the anaerobic bacterial blend or a placebo. After incubation, pro-inflammatory IL-8 production in the HT-29 cell line will be measured using ELISA assay. Later, the capability of a single freeze-dried strain in attenuating the IL-8 levels produced by HT-29 cell line will be assessed.
Caco-2 cell line will be used to measure the effect of the microbiota samples collected from M-SHIME(R) on the barrier integrity of the Caco-2 (enterocyte) monolayer. The integrity will be assessed by measuring the TEER (transepithelial/transendothelial electrical resistance) of the cell monolayer, and the results will tell us if our strains can alleviate leakiness in the epithelial cell layer caused by dysbiosis.

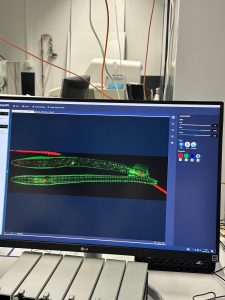
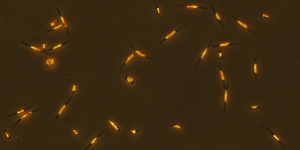
HT-29 cells under a microscope.
Starting my research
So far, I’ve read lots of literature related to the gut microbiota and treating inflammation with probiotics and next-generation probiotics. This has given me a good foundation on which to build my knowledge. The steps using M-SHIME(R) human gut simulator have been completed previously, so I’ve mainly been growing the intestinal cell lines required for our experiments. Luckily, I already have some experience with cell culture, so the start hasn’t been too difficult. My group has also been supportive and helpful, so I’m optimistic about the upcoming months!
Seeding HT-29 cells for our experiments.
The next step will be starting the IL-8 induction experiments, and once the initial results are ready, we can proceed to plan further. If you’re interested in our results, stay tuned for my next blog post!
Sources:
Bastos AR, Pereira-Marques J, Ferreira RM, Figueiredo C. Harnessing the Microbiome to Reduce Pancreatic Cancer Burden. Cancers (Basel). 2023 May 5;15(9):2629. doi: 10.3390/cancers15092629. PMID: 37174095; PMCID: PMC10177253.
Kouzu K, Tsujimoto H, Kishi Y, Ueno H, Shinomiya N. Bacterial Translocation in Gastrointestinal Cancers and Cancer Treatment. Biomedicines. 2022 Feb 4;10(2):380. doi: 10.3390/biomedicines10020380. PMID: 35203589; PMCID: PMC8962358.
Martín R, Langella P. Emerging Health Concepts in the Probiotics Field: Streamlining the Definitions. Front Microbiol. 2019 May 21;10:1047. doi: 10.3389/fmicb.2019.01047. PMID: 31164874; PMCID: PMC6536656.
Some literature:
Berta Bosch, Saliha Moutaharrik, Andrea Gazzaniga, Kaisa Hiippala, Hélder A. Santos, Alessandra Maroni, Reetta Satokari. Development of a time-dependent oral colon delivery system of anaerobic Odoribacter splanchnicus for bacteriotherapy. European Journal of Pharmaceutics and Biopharmaceutics. 2023; 190. 73-80. https://doi.org/10.1016/j.ejpb.2023.07.010.
Cristofori F, Dargenio VN, Dargenio C, Miniello VL, Barone M, Francavilla R. Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation: A Door to the Body. Front Immunol. 2021; 26(12):578386. doi: 10.3389/fimmu.2021.578386.
Marzorati M, Van den Abbeele P, Bubeck SS, Bayne T, Krishnan K, Young A, Mehta D, DeSouza A. Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in an M-SHIME® Model. Microorganisms. 2020; 8(7):1028. https://doi.org/10.3390/microorganisms8071028