What can you do when you are supposed to go to sea to sample plankton and you notice in the morning that a great amount of ice has floated in to block the harbour? Well, go slowly and carefully and prepare for a workout and some cold fingers. And how do the changes in phytoplankton communities affect the global methane budget? I guess that’s what we want to research.
Hi, I am Evert Odé, a second-year Bachelor’s student in Environmental Sciences at the University of Helsinki and one of the recipients of this year’s HiLIFE Research Trainee Scholarships. I am especially interested in marine ecosystems and their interactions with the Earth and climate systems in these agonising times of great human-induced changes. Phytoplankton, those tiny organisms at the base of most marine food webs, are at the centre of these interactions: they affect the climate system by sequestering carbon dioxide and influencing the formation of clouds but are also affected by climate change.
It is well known that phytoplankton constitute an important carbon sink, and people have even proposed that the oceans should be fertilised with iron to fight climate change. However, phytoplankton can also emit greenhouse gases. This all makes the interaction between phytoplankton and climate change a very fascinating subject indeed, and I am glad that I have the opportunity to conduct my four-month internship in the Plankton Biodiversity Research group at Tvärminne Zoological Station and participate in their research on this matter.

Me and a nice view of the archipelago off Tvärminne in early March
One topic we are especially interested in studying is methane production by different kinds of phytoplankton. For a long time, it has been a paradigm that biotic methane production only happens in environments where oxygen is not present1, such as sediments and bogs rich in decaying organic matter, and the digestive systems of ruminant animals. Moreover, only certain types of Archaea, those intriguing members of the third domain of life, have been considered capable of methane production2: the methanogens, which inhabit these anoxic environments. However, there have been signs that this standpoint probably doesn’t explain all methane production that is being detected. Especially the phenomenon called the “marine methane paradox” has been challenging to explain from this point of view: methane concentrations in oceanic surface waters are in supersaturation relative to the atmosphere (that is, they contain more methane than they would if they would be in balance with the atmospheric concentrations), even though they are rich in oxygen and far away from the potentially anoxic sediments1.
In recent decades, the paradigm of methanogens as the only methane producers has begun to crumble, as an accumulating number of studies have revealed that many organisms in both Eukaryote and Bacteria domains do produce methane, even though the exact pathways are not always clear. For us, the most interesting aspect of this new research is that a number of phytoplankton species have been demonstrated to produce methane, and even in association with photosynthesis, which produces oxygen! Since oxygen (a very oxidising substance, as could be concluded from the name) could be considered an enemy of methane, a reduced hydrocarbon substance that has been thought to form only in anoxic conditions, this is quite a counterintuitive finding. It also illustrates that one should be cautious with paradigms, especially in the field of microbiology.
The most important aspect of all this might be how these new findings affect the global methane budget. Even though the methane production by phytoplankton and other organisms is a natural phenomenon that has been going on for millions of years, it can significantly affect the calculations of global emissions and sinks of methane3. This could, in turn, affect our thoughts concerning significance and quantification of other, more human-influenced sources of this potent greenhouse gas. In addition, phytoplankton-derived methane emissions can also change notably4 due to the great anthropogenic changes of our time.
The phytoplankton community consists of many groups of organisms with very distinct evolutionary backgrounds and widely differing traits. Cyanobacteria, those ancient blue-greenish organisms that have the honour to be the evolutionary innovators of oxygenic photosynthesis, have been shown to produce methane2. Their summer blooms have become a common phenomenon in the eutrophicated and warming Baltic Sea, as well as in many lakes (a well-known nuisance to many enjoyers of aquatic environments), and in our project, we want to further investigate the climatic effects of their increasing dominance. Their competitors among the summer phytoplankton include the cryptophytes which we are also interested in researching.

Starting to inoculate some algae for our experiments (in this case Synechococcus sp., very small cyanobacteria)
Probably an even more interesting group of phytoplankton from our point of view are the dinoflagellates. They are indeed strange creatures: many of them photosynthesize with chloroplasts acquired through secondary, tertiary and even quaternary endosymbiosis (which means that their ancestors have engulfed an alga, whose ancestors have engulfed an alga, whose ancestors have engulfed an alga, whose ancestors have engulfed a cyanobacterium… crazy, isn’t it?), but they can also eat other organisms. And move around the sea by whirling their flagella!
Methane production by dinoflagellates has not been researched much, even though they are increasing in abundance in many areas, including the Baltic Sea. Their increase here during the phytoplankton spring bloom has come at the expense of diatoms. Diatoms are another, still more common and diverse group of phytoplankton. They do not eat anybody or move around much, but they are quite efficient in photosynthesis and nutrient uptake, and their surface is made of glass (or very nearly so). In the upcoming experiment this spring, we will investigate the effects of the dinoflagellates, diatoms, cyanobacteria and cryptophytes on the sequestration and release of methane and carbon dioxide.
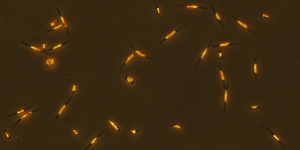
Cells of Phaeodactylum tricornutum, a diatom species that we are using in our experiment, with two different morphological types visible in the picture
Well, what about the struggle against the ice floes? In addition to laboratory experiments, our project also includes field monitoring and measurements. The ice situation at the beginning of March was difficult due to the fast ice surrounding the harbour. When the fast ice finally broke, we decided to sample on next Thursday morning. On Wednesday, I watched from the window of my microscopying room as the countless ice floes floated slowly from the sea to the harbour driven by some change of wind. It didn’t seem very promising for our field work.
The next morning, the drift ice was still there, blocking the harbour, even though the waters further out were quite ice-free. We still obtained a permit to try; but very slowly… So, we boarded Crangon, an 8,2–metre–long research vessel, and started a long and arduous journey through the drift ice. Research technician Kurt Spencer drove the boat, while postdoc researcher Per Hedberg kept the ice floes away from the propeller with a boathook and I tried to push ice aside from our route at the front of the boat. It took an hour for us to reach the open water not so far away, but we were very satisfied when finally getting to take our samples. This was done quite quickly and then… all the way back again. Hopefully this would not take much longer, for then we would miss lunch.

The drift ice that we had to struggle through (the weather was foggy, which added nicely to the ambience of the scene).
So, we started pushing ice diligently once again. I was unsatisfied with my telescopic boathook pole, which clearly was not designed to be used for this purpose and didn’t stay in its length when pushing the ice floes away. When we started returning to the harbour through drift ice, I saw a long piece of driftwood, probably a fugitive lath from some faraway building site. I picked it up from the icy sea with the boathook, and it turned out to be a useful tool for ice manipulation. Sometimes, driving through a combination of many large ice floes was quite a puzzle, but in the end, we managed to make it back by lunch. Even though my fingers were quite cold from sampling and handling the wet piece of wood, I thought it was a fun way of doing science.
Some literature:
1Bižić, M., Grossart, H.-P. and Ionescu, D. (2020) Methane Paradox, In eLS, John Wiley & Sons, Ltd (Ed.). https://doi.o rg/10.1002/9780470015902.a0028892.
2Bižić, M. et al. (2020) Aquatic and terrestrial cyanobacteria produce methane. Science advances. [Online] 6 (3), eaax5343–eaax5343.
3Günthel, M. et al. (2019) Contribution of oxic methane production to surface methane emission in lakes and its global importance. Nature communications. [Online] 10 (1), 5497–10.
4Klintzsch, T. et al. (2020) Effects of Temperature and Light on Methane Production of Widespread Marine Phytoplankton. Journal of geophysical research. Biogeosciences. [Online] 125 (9).