Transfer RNAs (tRNAs) are often described as humble, clover shaped molecular servants. They participate in protein synthesis by performing codon recognition on messenger RNAs (mRNAs) and by delivering amino acids necessary for translation. Despite technically being an accurate description, it underestimates the profound intricacies of the tRNAs. Humans have around 600 tRNA genes in the genome, and when expressed and processed, the beautiful clover leaf tRNA is reshaped to an L-shaped configuration and will acquire variable chemical modifications, increasing the diversity of tRNAs (Lant et al., 2019). Altogether, the modified L-shaped tRNAs comprise 15% of the total RNA found in the cell, whilst mRNA only comprises 1-5 % (Delaunay et al., 2024). Disruptions in tRNA expression, regulation and mutations have been linked with neurological and metabolic disorders and cancer (Lant et al., 2019). tRNAs are found in all forms of life (Delaunay et al., 2024), underscoring their fundamental role in biology. Understanding tRNAs is essential for understanding life: after all, would life even exist without the humble tRNA?
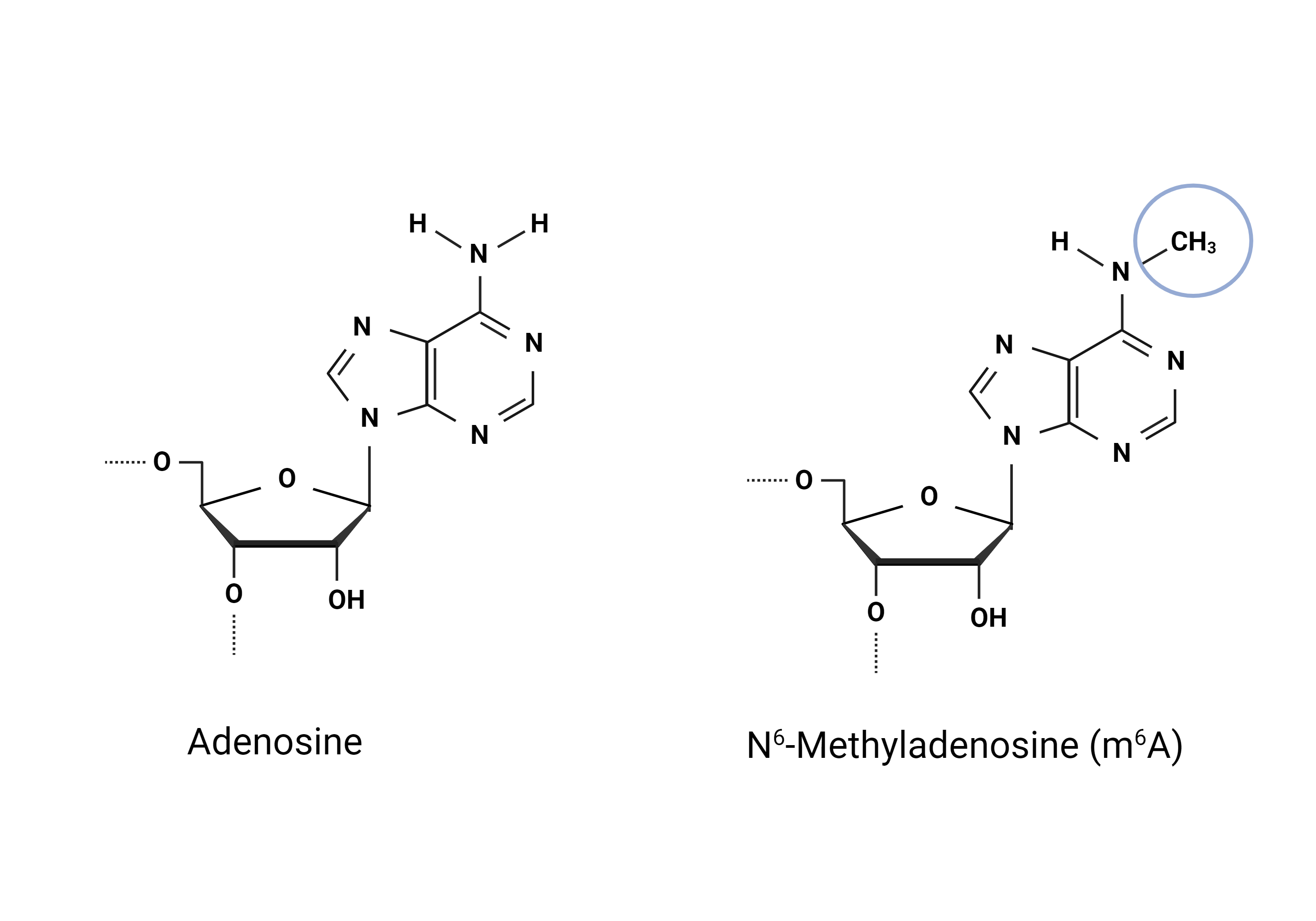
Modification of cellular macromolecules is crucial for accurate and efficient gene regulation. Many are familiar with DNA modifications, such as cytosine methylation, which may affect gene activity and chromatin structure (Liyanage et al., 2014). Cellular RNAs are also targets of post-transcriptional modifications (PTMs) with the N6-Methyladenosine (m6A) modification being one of the most widely studied (Lauman & Garcia, 2020). RNA modifications have essential regulatory implications: in mRNAs, certain modifications can affect transcript stability, localisation, splicing patterns, and translation (Delaunay et al., 2024). PTMs can be found in ribosomal RNA, long non-coding RNAs, and small non-coding RNAs (Delaunay et al., 2024). Unsurprisingly, PTMs are also seen in tRNAs. In fact, tRNAs are the most abundantly modified RNA species in the cell (Zhang et al., 2022). Our understanding of their effects is still limited but we know that the modifications can affect tRNA stability, tRNA-RNA interactions, tRNA-protein interactions, folding and mRNA decoding (Delaunay et al., 2024). Over 150 tRNA modifications have been identified (Delaunay et al., 2024) and as tRNAs are abundant in the cells, studying tRNA modifications becomes a very intriguing area of research.
During my time as a HiLIFE trainee I had the opportunity to delve into the science of tRNA modifications at the RNAcious laboratory, University of Helsinki. I participated in two projects, one where the aim was to produce hypomodified tRNAs and another where the aim was to determine the tRNA modification landscapes in different mouse tissues. You can find my first blog post here: https://blogs.helsinki.fi/hilife-trainees/2023/06/26/my-battle-against-rnases/
The making of plain cloverleaves: Hypomodified tRNAs
Modifications on tRNAs are so abundant that it would be difficult, or maybe even impossible, to extract hypomodified tRNAs from the cell: you see, in eukaryotes there are on average 13 modifications on each ~80nt long tRNA (Zhang et al., 2022). For methylations alone, there are around 40 proteins, known as modification writers (e.g. methyltransferases) and erasers (e.g. demethylases), which moderate tRNA modifications (Delaunay et al., 2024). One way to produce hypomodified tRNAs is through in vitro transcription (IVT), which essentially is a cell free transcription system. As IVT is cell free, it lacks the writer and eraser enzymes which modify the modification profile. My task was to develop a method to produce and isolate hypomodified tRNAs utilizing IVT, ribozyme- and MS2 based techniques.
Differentially decorated cloverleaves: tRNA modifications in different organs
We know that there are different tRNA modification landscapes in different organs (de Crécy-Lagard et al., 2019) but they have not yet been studied extensively. A tRNA modification landscape is the entire modification profile of the tRNAs of a specific organ. Uncovering the modification landscape would offer valuable insights into both the frequency and positional distribution of specific modifications within the tRNAs across various organs. Mass spectrometry is an effective tool to identify and study the location of modifications on single nucleotides (Lauman & Garcia, 2020), and certain reverse transcriptases can be used to study the location of modifications on the tRNA molecule (Zhang et al., 2022).
After my time as a HiLIFE trainee, I’ve truly gained a deep appreciation for the complexities of the humble tRNA. While I metaphorically refer to tRNA modifications as “decorations”, in reality, these modifications play essential roles in biological processes. Learning about them has been truly fascinating.
To end this journey, I would like to express my gratitude to Docent Peter Sarin and his group of bright researchers, especially my supervisor Jenni Pedor, who always supported me during my time in the research laboratory. The pioneering and motivational environment provided an invaluable experience and an inspiration for my career moving forward. I would encourage anyone interested in expanding their understanding of tRNA modifications and tRNA biology to explore the research conducted at RNAcious laboratory.
References
de Crécy-Lagard, V., Boccaletto, P., Mangleburg, C. G., Sharma, P., Lowe, T. M., Leidel, S. A., & Bujnicki, J. M. (2019). Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic acids research, 47(5), 2143–2159. https://doi.org/10.1093/nar/gkz011
Delaunay, S., Helm, M., & Frye, M. (2024). RNA modifications in physiology and disease: towards clinical applications. Nature reviews. Genetics, 25(2), 104–122. https://doi.org/10.1038/s41576-023-00645-2
Lant, J. T., Berg, M. D., Heinemann, I. U., Brandl, C. J., & O’Donoghue, P. (2019). Pathways to disease from natural variations in human cytoplasmic tRNAs. The Journal of biological chemistry, 294(14), 5294–5308. https://doi.org/10.1074/jbc.REV118.002982
Lauman, R., & Garcia, B. A. (2020). Unraveling the RNA modification code with mass spectrometry. Molecular omics, 16(4), 305–315. https://doi.org/10.1039/c8mo00247a
Liyanage, V. R., Jarmasz, J. S., Murugeshan, N., Del Bigio, M. R., Rastegar, M., & Davie, J. R. (2014). DNA modifications: function and applications in normal and disease States. Biology, 3(4), 670–723. https://doi.org/10.3390/biology3040670
Zhang, W., Foo, M., Eren, A. M., & Pan, T. (2022). tRNA modification dynamics from individual organisms to metaepitranscriptomics of microbiomes. Molecular cell, 82(5), 891–906. https://doi.org/10.1016/j.molcel.2021.12.007